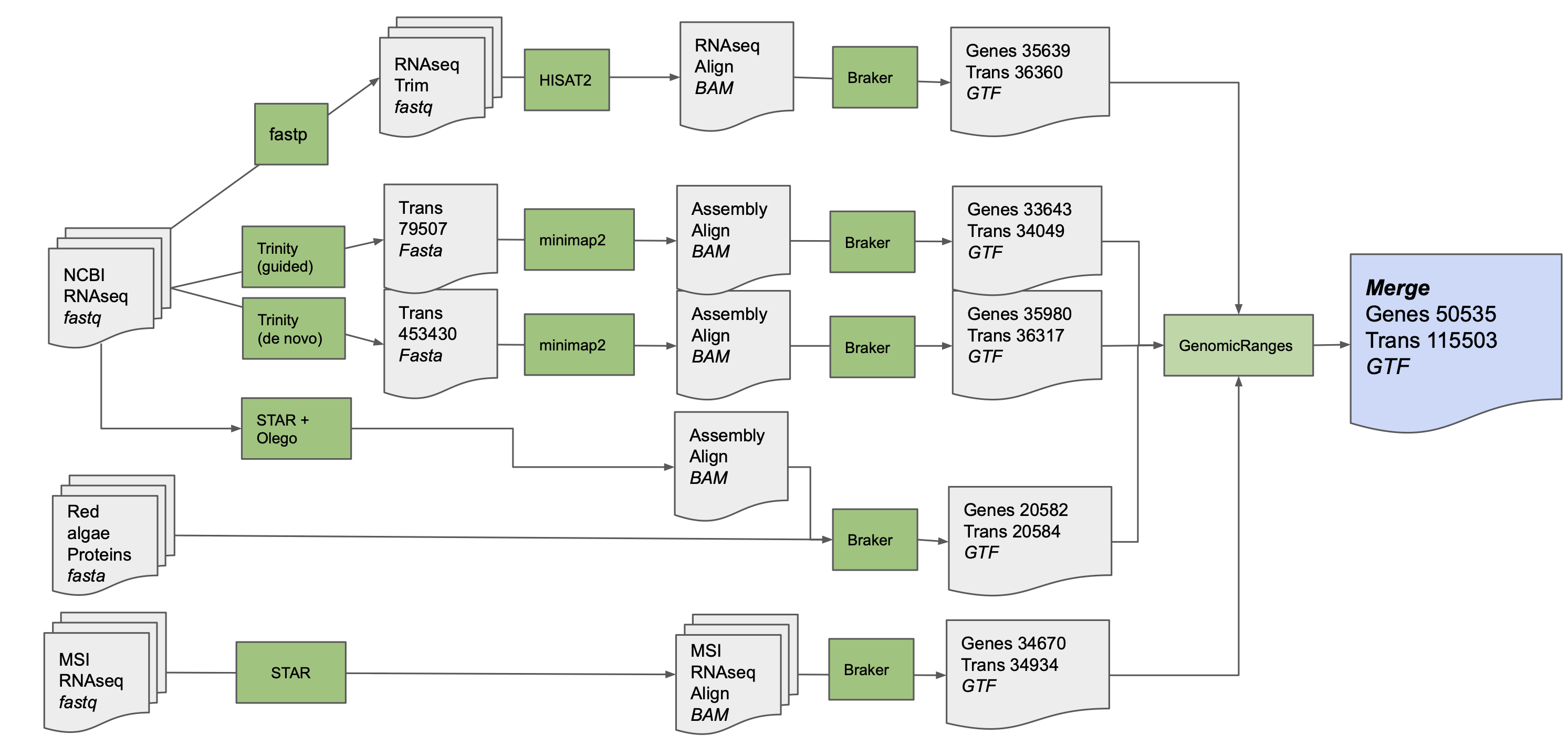
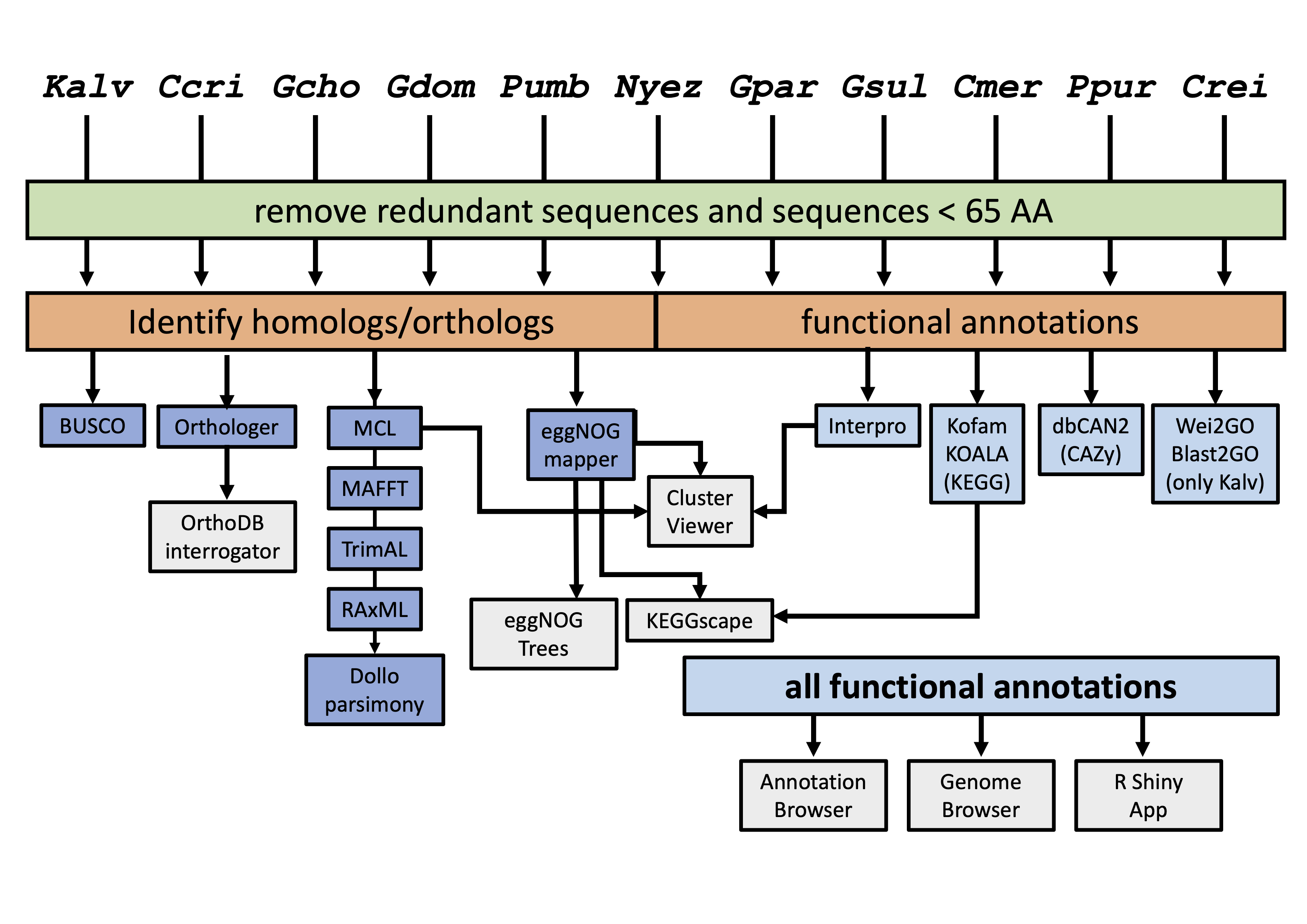
KaGE: Kappaphycus alvarezii Genome Explorer
Welcome to KaGE - the Kappaphycus alvarezii Genome Explorer.
Use this platform to browse the complete, assembled genome of Kappaphycus alvarezii,
accompanying annotations and perform basic analyses on this data.
To cite KaGE resources please refer to:
The Kappaphycus alvarezii Genome Explorer (KaGE): Bioinformatics Resources for a globally cultivated seaweed" (Fahrenkrug SC, Crisostomo B, and Roleda M, 2023)